ReMap 2020: Atlas of Regulatory Regions
Description ReMap 2020 Homo Sapiens
This track hub represents the ReMap Atlas of regulatory regions which consists in a large scale integrative analysis of all Public ChIP-seq data for transcriptional regulators from GEO, ArrayExpress and ENCODE.
To enable genome-wide identification of regulatory elements we have collected, curated, analysed a total of 6,498 ChIP-seq and Exo-seq data sets from Public source (GEO, ArrayExpress and ENCODE). After applying our quality filters we retained 6,000 data sets.
Those merged analyses covers a total of 960 DNA-binding protein (transcriptional regulators) such as a variety of transcription factors (TFs), transcription co-activators (TCFs) and chromatin-remodeling factors (CRFs) for 164 million peaks.
Below a schematic diagram of the types of regulatory regions:
- ReMap 2020 Atlas
- ReMap 2020 Cis Regulatory Regions
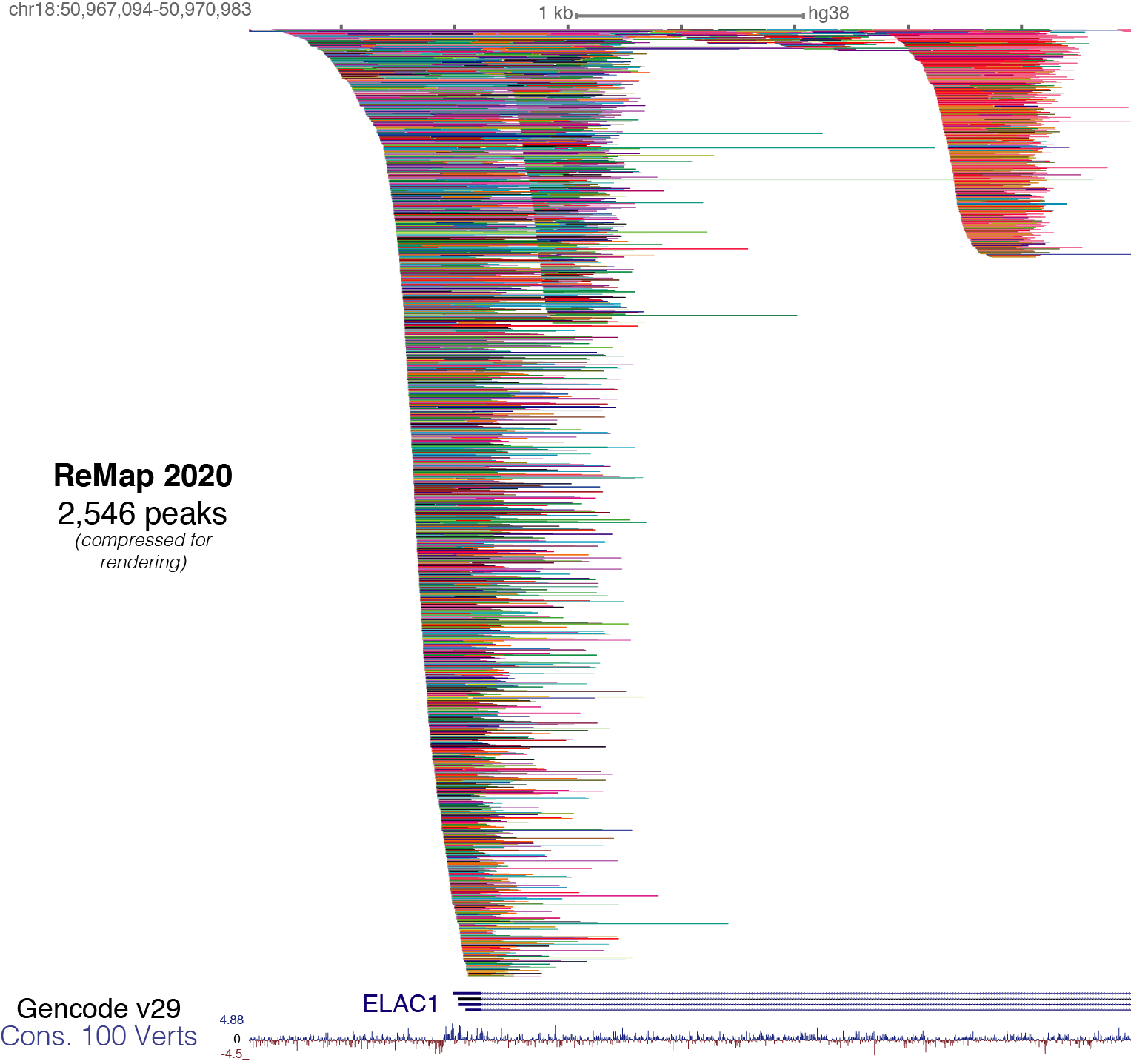
Individual BED files for specific TFs, or Cells or datasets can be found and downloaded on the ReMap website http://remap.cisreg.eu or http://remap.univ-amu.fr/
Description ReMap 2020 Arabidopsis thaliana
This track hub represents the ReMap Atlas of regulatory regions which consists in a large scale integrative analysis of all Public ChIP-seq data (and DAP-seq) for transcriptional regulators from NCBI-GEO.
We analysed 796 quality controlled ChIP-seq/DAP-seq experiments from the GEO data wharehouse. Those ChIP-seq (n=179 TRs, n=287 Histones), DAP-seq (n=330) datasets have been mapped to the TAIR10 assembly. Here we define a “dataset” as a ChIP-seq experiment in a given series (e.g. GSE94486), for a given target (e.g. ARR1), in a particular biological condition (i.e. ecotype, tissue type, experimental conditions ; e.g. Col-0_seedling_3d-6BA-4h).
Datasets were labeled by concatenating these three pieces of information such as GSE94486.NR2C2.Col-0_seedling_3d-6BA-4h.
Below a schematic diagram of the types of regulatory regions availble at http://remap.univ-amu.fr/:
- ReMap 2020 Atlas
- ReMap 2020 Non redundant peaks
- ReMap 2020 Cis Regulatory Regions
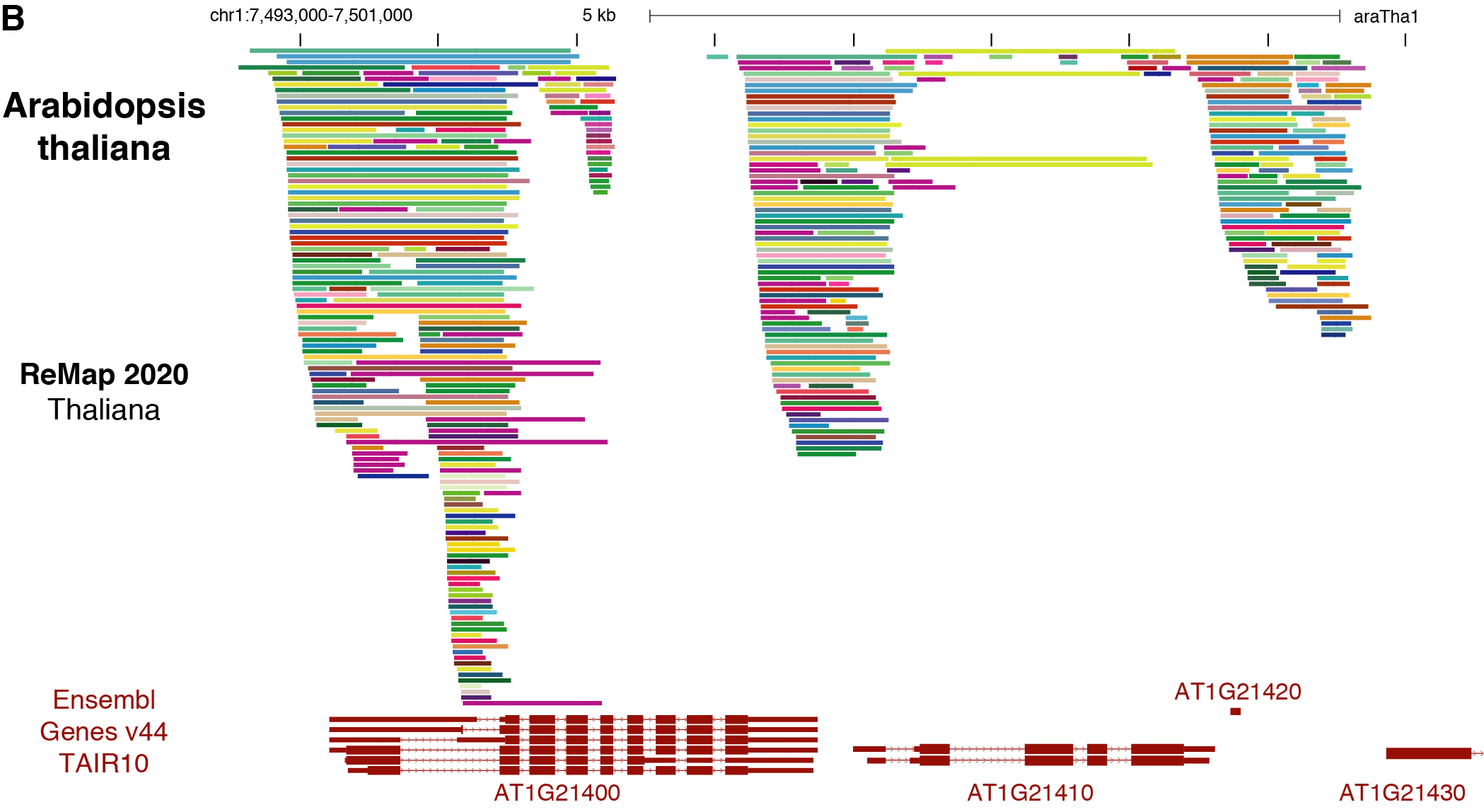
Individual BED files for specific TFs, or Cells or datasets can be found and downloaded on the ReMap website http://remap.cisreg.eu or http://remap.univ-amu.fr/
Display Conventions and Configuration
-
Each transcription factor follow a specific RGB color.
-
ChIP-seq peak summits are represented by vertical bars.
-
Hsap : A data set is defined as a ChIP/Exo-seq experiment in a given GEO/ArrayExpress/ENCODE series (e.g. GSE41561), for a given TF (e.g.: ESR1), in a particular biological condition (e.g. MCF-7).
Data sets are labelled with the concatenation of these three pieces of information (e.g. GSE41561.ESR1.MCF-7).
-
Atha : we define a “dataset” as a ChIP-seq experiment in a given series (e.g. GSE94486), for a given target (e.g. ARR1), in a particular biological condition (i.e. ecotype, tissue type, experimental conditions ; e.g. Col-0_seedling_3d-6BA-4h).
Data sets are labelled with the concatenation of these three pieces of information (e.g. GSE94486.ARR1.Col-0_seedling_3d-6BA-4h).
Methods
Human Data sets sources
GEO & ArrayExpress
Public ChIP-seq data sets were extracted from Gene Expression Omnibus (GEO) and ArrayExpress (AE) databases. For GEO, the query '('chip seq' OR 'chipseq' OR 'chip sequencing') AND 'Genome binding/occupancy profiling by high throughput sequencing' AND 'homo sapiens'[organism] AND NOT 'ENCODE'[project]' was used to return a list of all potential data sets to analyse, which were then manually assessed for further analyses. Data sets involving polymerases (i.e. Pol2 and Pol3), and some mutated or fused TFs (e.g. KAP1 N/C terminal mutation, GSE27929) were exckuded.
ENCODE
Available ENCODE ChIP-seq data sets for transcriptional regulators from www.encodeproject.org portal were processed with the our uniform workflow. We retrieved the list of ENCODE data as FASTQ files from the ENCODE portal (https://www.encodeproject.org/) using the following filters: Assay: "ChIP-seq", Organism: "Homo sapiens", Target of assay: "transcription factor", Available data: "fastq" on 2016 June 21st. Metadata information in JSON format and FASTQ files were retrieved using the Python requests module.
Human ChIP-seq processing
Both Public and ENCODE data were processed similarly. Bowtie 2 (PMC3322381) (version 2.2.9) with options -end-to-end -sensitive was used to align all reads on the human genome (GRCh38/hg38 assembly). Biological and technical replicates for each unique combination of GSE/TF/Cell type or Biological condition were used for peak calling. TFBS were identified using MACS2 peak-calling tool (PMC3120977) (version 2.1.1.2) in order to follow ENCODE ChIP-seq guidelines, with stringent thresholds (MACS2 default thresholds, p-value: 1e-5). An input data set was used when available.
Arabidopsis Data sets sources
After consistent peak calling across datasets, we identified 1.9 million peaks bound by transcriptionnal regulators from ChIP-seq data and 0.8 million from DAP-seq data (GSE60141), giving a regulatory atlas of 2.6 million peaks. These numbers may include overlapping sites for identical TR targets which were studied in various conditions. To address this we merged overlapping TRs binding regions for similar TRs obtaining a catalogue of 1.8 million non-redundant binding sites.
Finally we also applied our pipeline to all available histone ChIP-seq data and identifed 4.5 million broad and gapped peaks.
Arabidopsis ChIP-seq processing
Both ChIP-seq and DAP-seq data were processed similarly. Bowtie 2 (PMC3322381) (version 2.2.9) with options -end-to-end -sensitive was used to align all reads on the human genome (GRCh38/hg38 assembly). Biological and technical replicates for each unique combination of GSE/TF/Cell type or Biological condition were used for peak calling. TFBS were identified using MACS2 peak-calling tool (PMC3120977) (version 2.1.1.2) in order to follow ENCODE ChIP-seq guidelines, with stringent thresholds (MACS2 default thresholds, p-value: 1e-5). An input data set was used when available.
Human & Arabidopsis Quality assessment
To assess the quality of public data sets, we computed a score based on the cross-correlation and the FRiP (fraction of reads in peaks) metrics developed by the ENCODE Consortium (http://genome.ucsc.edu/ENCODE/qualityMetrics.html). Two thresholds were defined for each of the two cross-correlation ratios (NSC, normalized strand coefficient: 1.05 and 1.10; RSC, relative strand coefficient: 0.8 and 1.0). Detailed descriptions of the ENCODE quality coefficients can be found at http://genome.ucsc.edu/ENCODE/qualityMetrics.html. We used the phantompeak tools suite (https://code.google.com/p/phantompeakqualtools/) to compute RSC and NSC.
Full details of our methods can be found in the references below.
Data Availability
The ReMap BED files are available for download at the ReMap website http://remap.cisreg.eu or http://remap.univ-amu.fr/ in the download tab.
Papers to cite
-
Cheneby J., Menetrier Z., Mestdagh M., Rosnet T., Douida A., Rhalloussi W., Bergon A., Lopez F., Ballester B.
ReMap 2020: A database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments.
Nucleic Acids Research (2020) gkz945 https://doi.org/10.1093/nar/gkz945.
-
Cheneby J., Gheorghe M., Artufel M., Mathelier A., Ballester, B.
ReMap 2018: An updated regulatory regions atlas from an integrative analysis of DNA-binding ChIP-seq experiments.
Nucleic Acids Research (2018) gkx1092 https://doi.org/10.1093/nar/gkx1092.
-
Griffon A., Barbier Q., Dalino J., van Helden J., Spicuglia S., Ballester B.
Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape.
Nucleic Acids Research (2015) 43 (4): e27.
Contact
If you have questions or comments, please write to:


