Exonic Enhancers Are a Widespread Class of Dual-Function Regulatory Elements
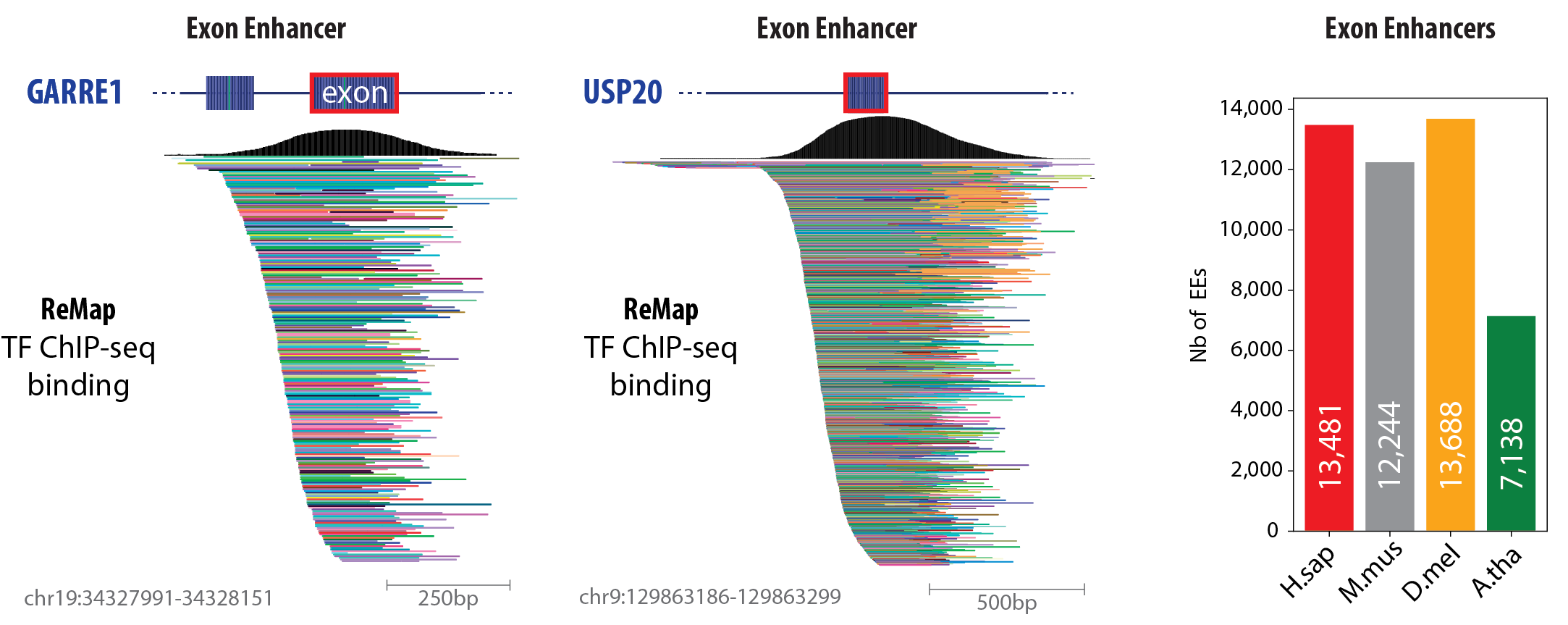 Two representative examples of EEs within protein-coding exons.
Two representative examples of EEs within protein-coding exons.
GARRE1 on human chromosome 19 and USP20 on chromosome 9.
The top black traces depict aggregated TF ChIP-seq signal (ReMap), illustrating a peak of transcription factor (TF) occupancy centered on an internal exon.
Each colored bar below represents a distinct TF dataset from ReMap, collectively underscoring extensive multi-TF binding at these exons.
Bar chart showing the number of EEs detected in four model organisms.
Description
This track hub represents the different Exonic Enhancers (EEs) identified by meta-analysis in four species.
Exonic enhancers (EEs) occupy an under-appreciated niche in gene regulation. By integrating transcription factor binding, chromatin accessibility, and high-throughput enhancer-reporter assays, we demonstrate that many protein-coding exons possess enhancer activity across species. These EEs exhibit characteristic epigenomic signatures, form long-range interactions with gene promoters, and can be disrupted by both nonsynonymous and synonymous variants.
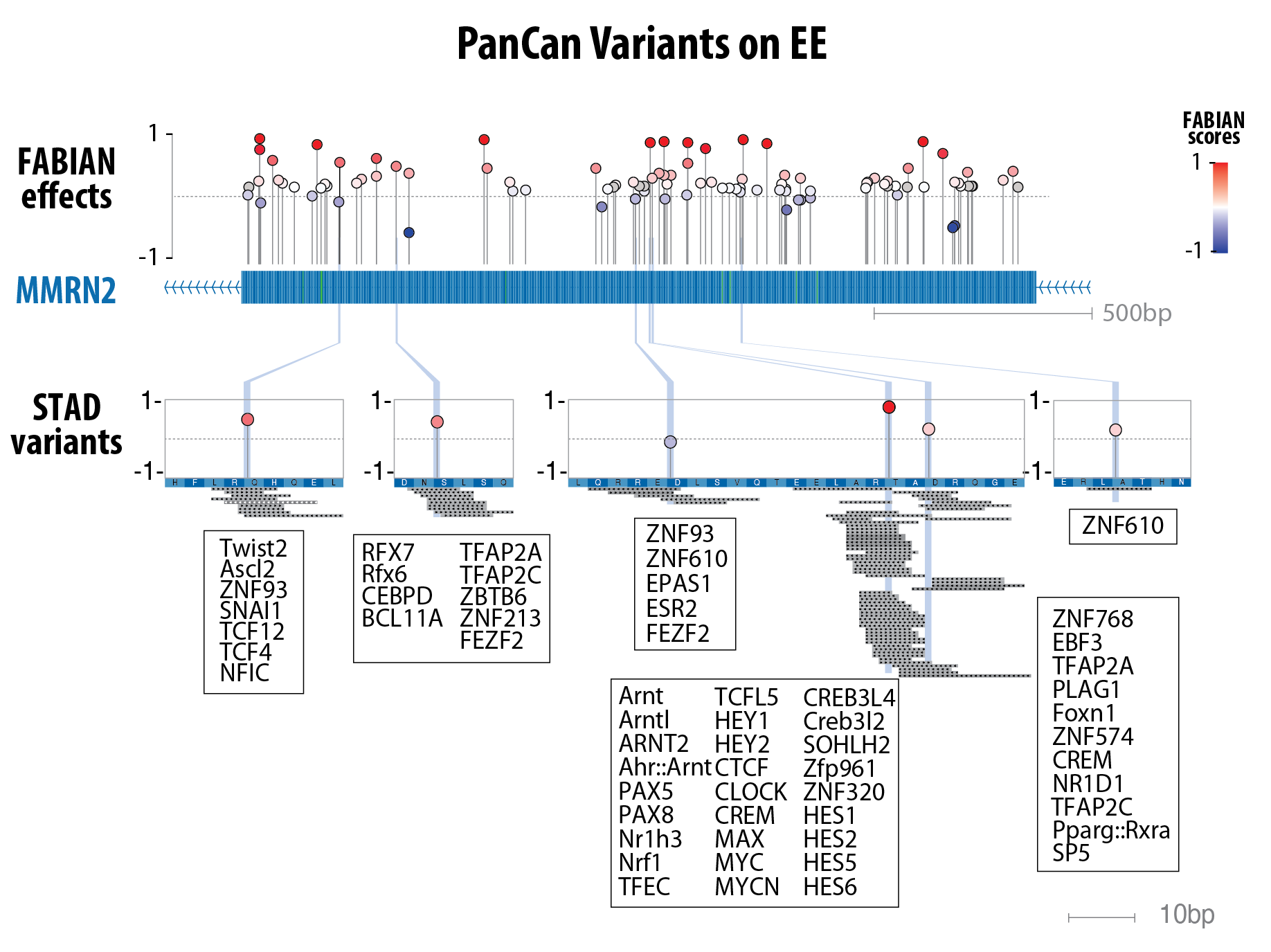 Lollipop plot representing PanCancerAtlas TF-binding disruptions in Exonic Enhancers.
Lollipop plot representing PanCancerAtlas TF-binding disruptions in Exonic Enhancers.
Example genomic track at the MMRN2 locus, illustrating predicted transcription factor binding changes from FABIAN variant analysis (4).
Red lollipops indicate variants that increase TF binding affinity, whereas blue lollipops denote decreased affinity.
The STAD (gastric cancer) variants align with multiple JASPAR TF motif sites (listed).
Data tracks in this hub
Human hg38
- EE track for hg38 assembly
- gnomAD variant effects on JASPAR TFBS matching ReMap ChIP-seq peaks
Human hg19
- EE track for hg19 assembly
- TCGA variant effects on JASPAR TFBS matching ReMap ChIP-seq peaks
- TCGA variant effects on JASPAR TFBS
Mouse mm39
- EE track for mm39 assembly
Arabidopsis araTha1
- EE track for araTha1 assembly
Variant Effects Predictions and Visualization in Exonic Enhancers
To assess how single-nucleotide polymorphisms (SNPs) may disrupt transcription factor (TF) binding within exonic enhancers (EEs), we used the
FABIAN-variant tool. FABIAN-variant quantifies binding affinity changes by evaluating the presence or absence of TF motifs at SNP-affected sites. For each variant (TCGA hg19, gnomAD hg38) located within an EE, we computed its predicted effect on associated TFBSs. Mutations that significantly altered TF-binding affinity (binding gain or loss) were recorded.
To facilitate the interpretation of TF-binding disruptions, we visualized the computed TF-binding impacts using lollipop representation in the UCSC Genome Browser. These visualizations provide a systematic overview of how mutations in exonic enhancers may alter TF-binding landscapes, potentially influencing gene regulation in cancer.
Data Availability
The Exonic Enhancers BED files for all species (Homo sapiens, Mus musculus, Drosophila melanogaster, Arabidopsis thaliana) are available in the supplementary data and the Zenodo repository of the paper.
Our study
-
Mouren JC., Torres M., van Ouwerkerk A., Manosalva I., Gallardo F., Spicuglia S., Ballester B.
Exonic enhancers are a widespread class of dual-function regulatory elements
In prep. (2025) .
Contact
If you have questions or comments, please write to:
References
-
Hammal F, de Langen P, Bergon A, Lopez F, Ballester B.
ReMap 2022: A database of regulatory regions from an integrative analysis of human and model organism transcription factor ChIP-seq experiments.
Nucleic Acids Research. 2022;50(D1):D316-D325.
ReMap Portal
-
Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, Santana-Garcia W, Tan G, Chèneby J, Ballester B, Sandelin A, Lenhard B, Wasserman WW, Mathelier A.
JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles.
Nucleic Acids Research. 2022;50(D1):D165–D173.
JASPAR Database
-
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al.
The mutational constraint spectrum quantified from variation in 141,456 humans.
Nature. 2020;581(7809):434–443.
Genome Aggregation Database (gnomAD)
-
Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, et al.; Cancer Genome Atlas Research Network.
Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer.
Cell. 2018;173(2):291-304.e6.
The Cancer Genome Atlas (TCGA)
-
Hombach D, Schuetzmann D, Lenz M, et al.
FABIAN-variant: Predicting transcription factor binding affinities for sequence variants of regulatory elements.
Nucleic Acids Research. 2022;50(W1):W322–W329.
FABIAN-variant Tool

