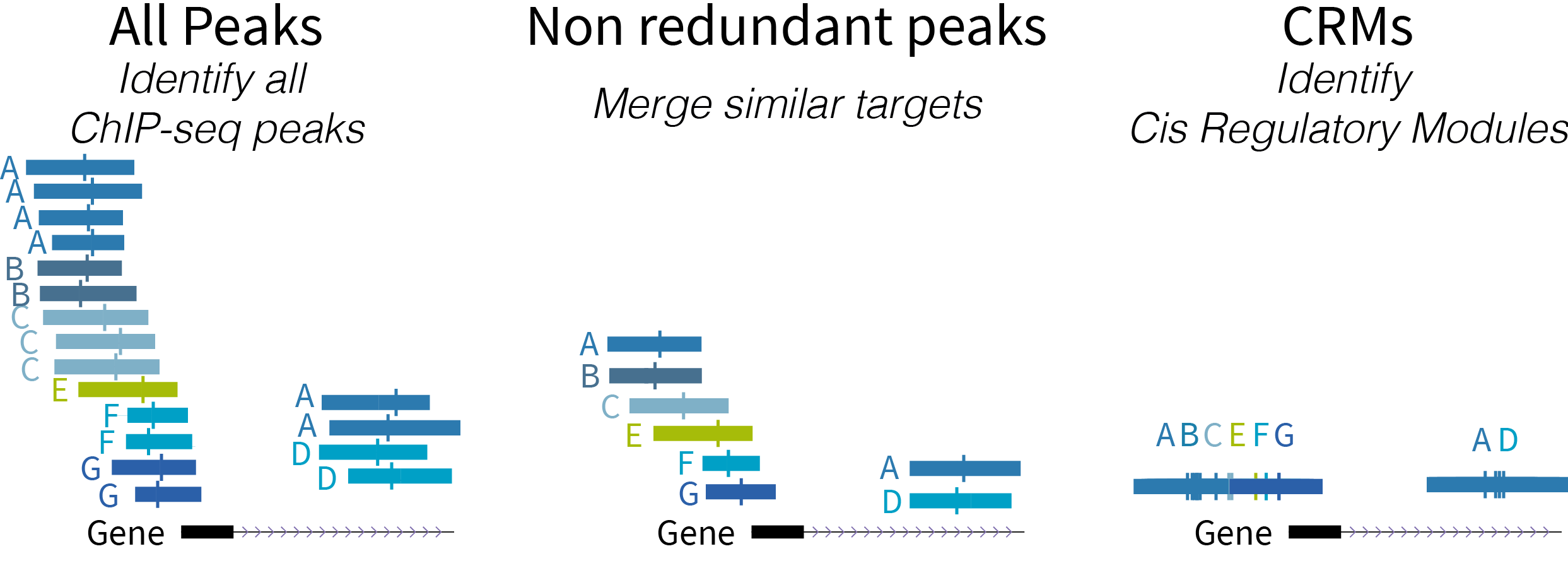
ReMap is a large scale integrative analysis of DNA-binding experiments for Homo sapiens, Mus musculus, Drosophila melanogaster and Arabidopsis thaliana transcriptional regulators. The catalogues are the results of the manual curation of ChIP-seq, ChIP-exo, DAP-seq from public sources (GEO, ENCODE, ENA).
ReMap database growth
Papers using ReMap
Long-read transcriptome sequencing reveals abundant promoter diversity in distinct molecular subtypes of gastric cancer. Huang KK, Huang J, Wu JKL, Lee M, Tay ST, Kumar V, Ramnarayanan K, Padmanabhan N, Xu C, Tan ALK, Chan C, Kappei D, Göke J, Tan P Genome biology 2021 Jan 22;22(1):44 10.1186/s13059-021-02261-x
Integrative pathway enrichment analysis of multivariate omics data. Paczkowska M, Barenboim J, Sintupisut N, Fox NS, Zhu H, Abd-Rabbo D, Mee MW, Boutros PC, Reimand J Nature communications 2020 Feb 5;11(1):735 10.1038/s41467-019-13983-9
Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer. Achinger-Kawecka J, Valdes-Mora F, Luu PL, Giles KA, Caldon CE, Qu W, Nair S, Soto S, Locke WJ, Yeo-Teh NS, Gould CM, Du Q, Smith GC, Ramos IR, Fernandez KF, Hoon DS, Gee JMW, Stirzaker C, Clark SJ Nature communications 2020 Jan 16;11(1):320 10.1038/s41467-019-14098-x
Mechanistic insights into transcription factor cooperativity and its impact on protein-phenotype interactions. Ibarra IL, Hollmann NM, Klaus B, Augsten S, Velten B, Hennig J, Zaugg JB Nature communications 2020 Jan 8;11(1):124 10.1038/s41467-019-13888-7