Species
Datasets sources
Statistics
| ChIP-seq | ChIP-exo | |
|---|---|---|
| Datasets (QC1+2 pass) | 7,895 | 208 |
| Targets | 1,002 | 208 |
| Peaks | 181,426,344 | 990,476 |
Integration of ChIP-seq and ChIP-exo data
In this ReMap 2022 human release we have manually curated and annotated 10,619 ChIP-seq experiments, retained after quality control 8,103 datasets, and in 2020 one large ChIP-exo experiment (GSE78099) were included from Imbeault M. et al Nature 2017. We applied our pipeline for both type of data, however most postprocessing steps applied by Imbeault M. et al Nature 2017 where applied to the ChIP-exo data. We processed another recent large ChIP-exo experiment (GSE151287, n=1259 datasets, from Lai W.K.M et al bioRxiv 2021) which was however not retained as the peak numbers identified after QCs were inconsistent with the publication.
After consistent peak calling and stringent QCs, we identified a total of 182 million peaks bound by transcriptionnal regulators (including 990,476 peaks from ChIP-exo). These numbers include overlapping sites for identical TRs which were studied in various conditions. To address this we merged overlapping TR peaks for similar TR obtaining a catalog of XX million non-redundant peaks.Datasets quality assessment
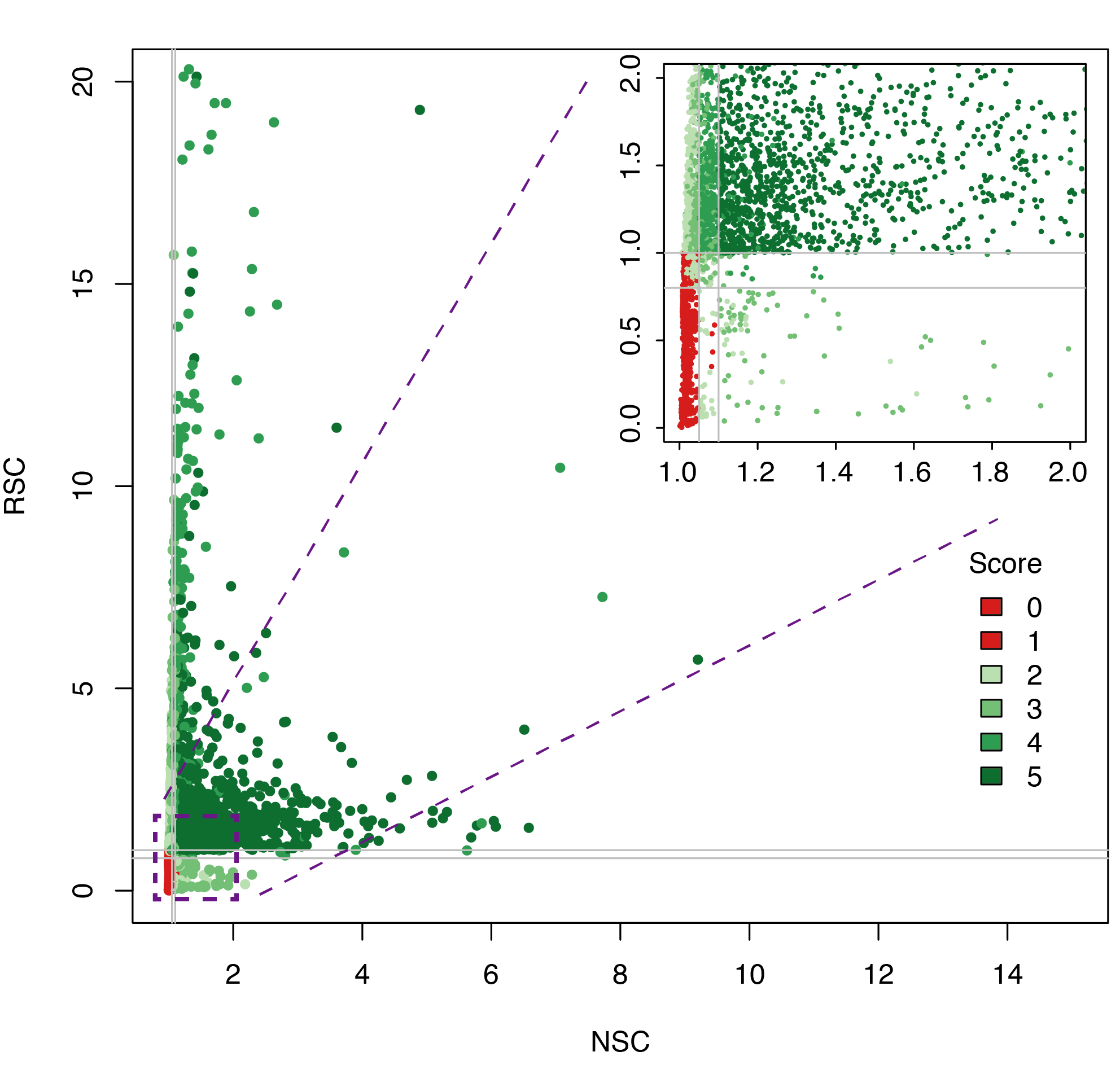
Datasets quality plot
ChIP-exo post-processing
Annotation and classification of transcription factors
Genomic visualization of peaks and analyses
A link to the UCSC Genome Browser was also added to facilitate genomic integration of the binding sites with other genome annotations. Our BED tracks allow for the visualization of our catalogues of binding sites on the human genome. Finally, different analyses such as the quality of datasets and DNA constraint analysis are provided for each transcription factor.
Downloading peaks
The ReMap BED files are available to download either for a given transcriptional regulator, by Biotype or for the entire catalog as one very large BED file.
For Homo sapiens the GRCh38/hg38 assembly is currently the supported assembly, but we lifted to hg19 with liftover. We provide archives of previous ReMap catalogs.
For Mus musculus we provide BED files for transcriptional regulators. The mm10 assembly is the assembly supported by ReMap, we probide lifted peaks in mm39.
For Drosophila melanogaster we provide BED files for transcriptional regulators. The dm6 assembly is the only assembly supported by ReMap.
For Arabidopsis thaliana we provide BED files for transcriptional regulators, histones marks, ecotypes and biotype coupled with a given ecotype. The TAIR10 assembly is the only assembly supported by ReMap.