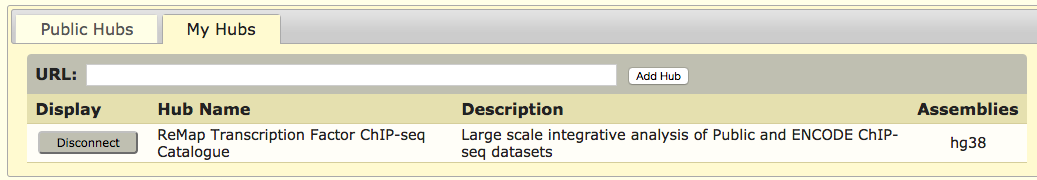
UCSC tracks
Direct link
All ReMap 2022 BED files have been uploaded and available as UCSC Genome Browser tracks, examples for Human in the EU or in the US servers.Users may want to directly browse the uploaded BEDs, as an alternative to Track Hub option, especially if users experience Track hub limitations on large regions.

Public session at UCSC
Sessions allow users to save snapshots of the Genome Browser and its current configuration, including the displayed tracks, position, and custom ReMap track data. The Public Sessions tool allows us to easily share our ReMap sessions that we find interesting with the rest of the community.
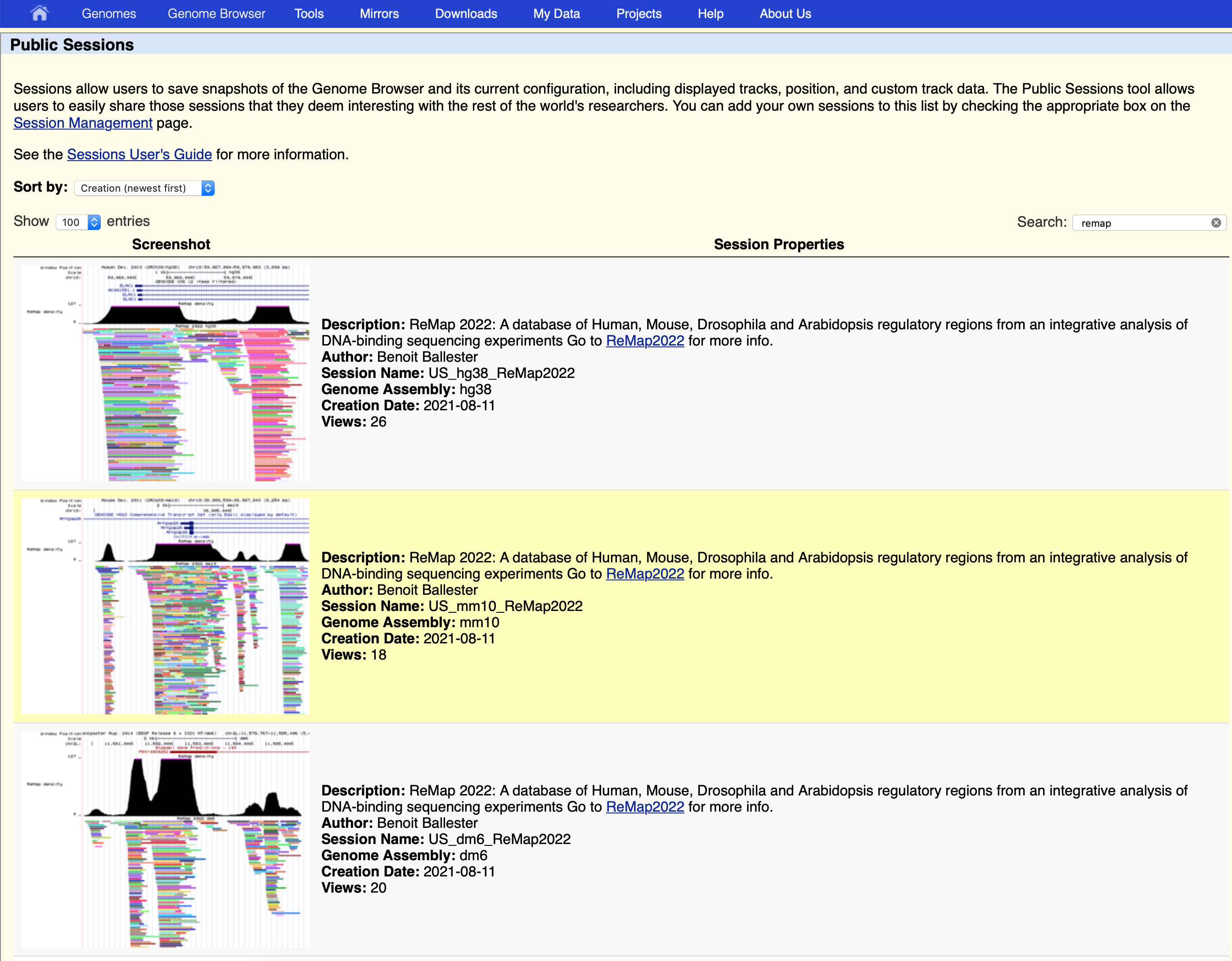
You can access public session directly https://genome.ucsc.edu/cgi-bin/hgPublicSessions .
Arabidopsis Histones Super Track at UCSC
We have created a super track to groups together related tracks such as histone tracks. By clicking the Histone link you’ll be able to switch on/off an of the track.
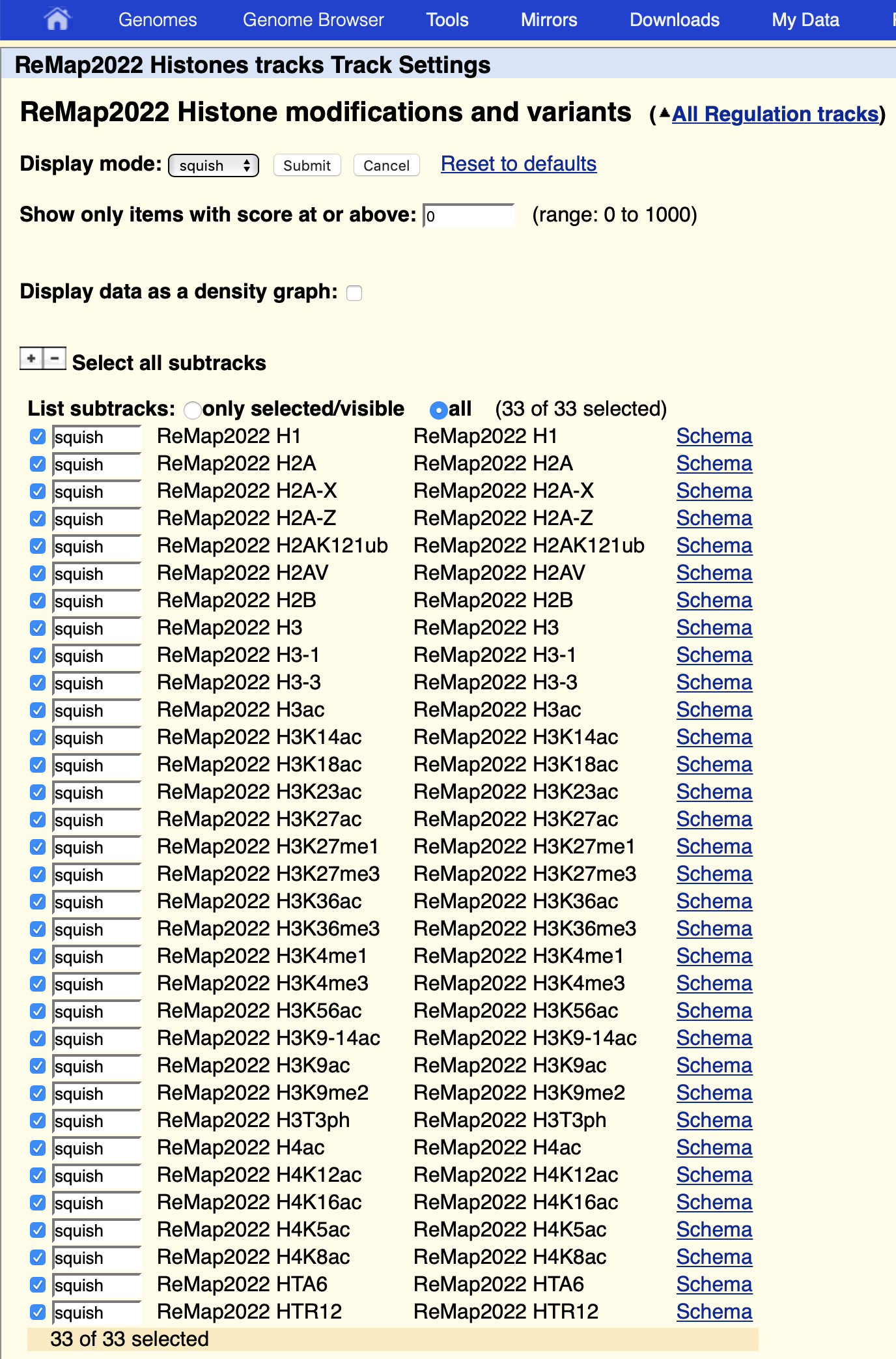
You can access histones super tracks via the UCSC track hub our public session directly https://genome.ucsc.edu/cgi-bin/hgPublicSessions . Contact us if you are having difficulties finding the supertrack configuration.
Ensembl tracks
Attach the ReMap track hub in e!
You can attach the ReMap track hub in Ensembl Human Location View, and it works very smoothly. Simply search for ReMap in the track hub repository, and connect the hub.
Alternatively you can add the this URL for ReMap2022http://remap.univ-amu.fr/storage/public/hubReMap2022/hub.txt.
For ReMap2020 you can use this URLhttp://remap.univ-amu.fr/storage/public/hubReMap2020UCSC/hub.txt.
For ReMap2018 you can use this URLhttp://remap.univ-amu.fr/storage/public/hubReMap2018/hub.txt.
Select track hub in e! tracks list
We advice to use the Stacked or Stacked unlimited track style for best display. Warning : Ensembl may limit the number of features diplayed in their browser, showing the message ""
Track Hub Registry
ReMap genome tracks are also available in Track Hub Registry. Use this link :
https://remap.univ-amu.fr/storage/public/hubReMap2022/hub.txt
New to Track Hubs ?
If you don't know what a Track Hub is, please check Using UCSC Genome Browser Track Hubs.
In short, Track hubs are web-accessible directories of genomic data that can be viewed on Genome Browsers. It offers a convenient way to view and share very large sets of data.
The track hub utility allows efficient access to data sets from around the world through the familiar Genome Browser interface. Users can display tracks from any Public Track Hubs that has been registered with UCSC.
Track Hub URL: https://remap.univ-amu.fr/storage/public/hubReMap2022/hub.txt
We strongly advice to check UCSC Track Hub paper, Raney BJ, et al. Track Data Hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014 Apr 1;30(7):1003-5.
Adding Track Hubs to Ensembl
The Ensembl team has put up a documentation page on How to attach a track hub to Ensembl.
If Ensembl is your favorite Genome Browser, follow this tutorial.
Due to the massive increase of data between ReMap 2014 and ReMap 2018, the Track Hub for the merged dataset (Public+ENCODE 80M peaks) may be displayed into Dense mode if the region being browsed is too large. This is because this particular track contains a lot of peaks. You may want to Zoom out, or switch off that track and keep both the Public and ENCODE tracks, or switch to Browsing the BED files from sessions directly, see below. This track is visible up to 50,000 peaks, then it switches to dense mode automatically.